2 What can you contribute and how?
Since 2000, OBIS has accepted, curated and published marine biodiversity data obtained by varied sources and methods. There is a common misconception that OBIS only accepts species occurrence data - however this is not true! OBIS can accept many types of marine data including:
- Presence/Absence
- Abundance, individual count
- Biomass
- Abiotic measurements
- Biotic measurements
- Sampling methods
- Sample processing methods
- Genetic data including sequences
- Data originating from historical records
- Tracking data
- Habitat data
- Acoustic data
- Imaging data
- Metadata describing the dataset and any project or programme related metadata
So if you have any of these types of marine data linked to your occurrence data and also want to contribute to OBIS - great! OBIS accepts data from any organization, consortium, project or individual who wants to contribute data. OBIS Data Sources are the authors, editors, and/or organisations that have published one or more datasets through OBIS. They remain the owners or custodians of the data, not OBIS!
OBIS harvests and publishes data from recognized IPTs from OBIS nodes or GBIF publishers. If you own data or have the right to publish data in OBIS, you can contact the OBIS secretariat or one of the OBIS nodes, or additionally a GBIF publisher. Your organization or programme can also become an OBIS node. An OBIS node usually publishes data from multiple data holders, effectively being a node in a network of data providers. So you may have to first find a relevant node before you get your data ready to publish.
To publish a dataset to OBIS, there are five main steps you must go through.
- First, you must identify which OBIS node is best suited to host your published data. If you would like to publish to GBIF at the same time, that is also possible. If your organization is already affiliated with a GBIF node with which you must publish from, OBIS can also harvest from GBIF nodes.
- Second, you must determine the structure of your data and which format will best suit your dataset. OBIS follows Darwin Core Archive (DwC-A) standards for datasets, and currently follows a star schema format. This format is based on relational databases. If you are unfamiliar with such database structures, or would like to refamiliarize yourself with them, please read here
- Then, you need to actually format your data according to OBIS and DwC-A standards and guidelines
- Once formatted, you should run a series of quality control measures to ensure you are not missing any required information and that all standards are being met. This helps ensure all data published in OBIS is formatted in a standardized way. When published in OBIS, OBIS provides a quality report to inform data owners and users of any quality control issues. By completing quality control before you publish your dataset you ensure there are fewer errors to fix later.
- Now that your dataset is ready for publishing, the relevant metadata must be filled in, and then published on the previously identified IPT.
Each of these steps are covered in detail in the relevant sections of the manual. For an overview of this process see data management flow in OBIS.
2.1 Why publish data to OBIS
It is important to publish and ensure your dataset follows a universal standard for several reasons. The FAIR guiding principles for scientific data management and stewardship provide a good framework to understand the reasoning behind publishing data. FAIR stands for Findable, Accessible, Interoperable, and Reusable. Let’s understand each aspect within the FAIR framework and how it is linked to publishing data in OBIS.
- F - Findable
Even if you publish your dataset on its own, publishing your data with OBIS will make your data more Findable (and Accessible) to a wider audience you might not have otherwise reached. By publishing your dataset to OBIS you are adding to a global database where your data can be found and analyzed alongside thousands of other datasets. For example, a dataset on marine invasive species in Venezuela was published July 20, 2022 and as of October 5, 2022 records of this dataset were included in 1,873 data download requests. This can save you time rather than handling individual data requests.
- A - Accessible
Similar to being Findable, OBIS makes your datasets more Accessible. Each dataset is given an identifier when you upload it on an IPT. Thus when users obtain data from OBIS, the original dataset can easily be identified and accessed. Data from OBIS is accessible in numerous ways, giving data users multiple avenues to potentially access your data.
- I - Interoperable
Using a standardized data format with controlled vocabularies will ensure your data are more Interoperable - more easily interpreted and processed by computers and humans alike. Increasingly, scientists use computer programs to conduct e-Science and collect data with algorithms. Formatting your data for OBIS will ensure it can be read and accessed by such programs as well as understood by users.
- R - Reusable
Publishing your data allows it to be Reused according to your chosen data usage license. Very likely you expended resources to collect your data and it would be a waste of those resources to leave your unique data unpublished and inaccessible for current and future generations. Likewise, it is better to preserve any data processing done to ensure your dataset is reproducible and/or verifiable. Finally, data in OBIS is often used in several assessment processes and used as information to support policy makers around the globe making informed decisions.
There are many other benefits of publishing in OBIS, even if you haven’t published any work on it yet. This includes:
- Your dataset can be associated with a DOI, allowing for your dataset to be more easily cited. By ensuring your dataset citation is complete you will ensure you are being cited properly.
- Publishing your dataset with OBIS makes it easier to set it up as a Data paper, which generates value for you and other researchers.
- There are social benefits to data publishing as your work becomes integrated into a wider dataset. It gives both you and your data more visibility. This can lead to more opportunities for collaboration and further career development as a researcher or professional.
- Your data can be incorporated into larger analyses to better understand global ocean biodiversity, helping to shape regional and international policies.
2.2 How to handle sensitive data
We recognize that sometimes your dataset may contain sensitive information (e.g., location data on endangered or poached species), or perhaps your organization does not want certain details publicly accessible. Types of sensitive data include:
- Location data on endangered or protected species
- Information regarding a commonly poached species
- Species or locations that have an economic impact (positive or negative)
To accommodate sensitivity but still be able to contribute to OBIS, we suggest:
- Generalizing location information by: Obtaining regional coordinates using MarineRegions, Getty Thesaurus of Geographic Names, or Google Maps
- Using the OBIS Map tool to generate a polygon area with a Well-Known Text (WKT) representation of the geometry to paste into the
footprintWKTfield. - Delay timing of publication (e.g., to accommodate mobile species)
- Submit your dataset, but mark it as private in the IPT so it is not published right away (i.e., until you set it as public). Alternatively, you can set a password on your dataset in order to share with specific individuals. Note that setting passwords will require some coordination with the IPT manager. By submitting your data to an IPT but not immediately publishing it, you can ensure that the dataset will be in a place to be incorporated at a later date when it is ready to be made public. This not only saves time and helps retain details while relatively fresh in your mind, but also ensures the dataset is still ready to be mobilized in case jobs are changed at a later date.
GBIF has created the following Best Practices for Generalizing Sensitive data which can provide you with additional guidance. Chapman AD (2020) Current Best Practices for Generalizing Sensitive Species Occurrence Data. Copenhagen: GBIF Secretariat. https://doi.org/10.15468/doc-5jp4-5g10.
2.3 OBIS Data Life Cycle
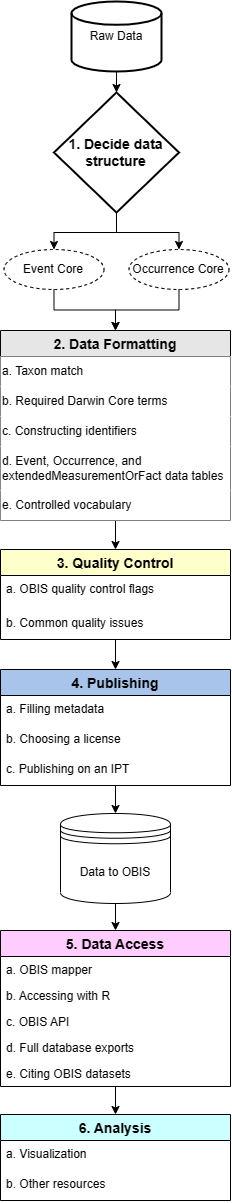
The basic data life cycle for contributions to OBIS can be broken down into six step-by-step phases:
- Data structure
- Data formatting
- Quality control
- Publishing
- Data access (downloading)
- Data visualization
Each of these phases are outlined in this manual and are composed of a number of steps which are covered in the relevant sections.
After you have decided on your data structure and have moved to the Data Formatting stage, you must first match the taxa in your dataset to a registered list. In formatting your dataset you will ensure the required OBIS terms and identifiers are mapped correctly to your data fields and records.
Depending on your data structure, you will then format data into a DwC-A format with the appropriate Core table (Event or Occurrence) with any applicable extension tables. Any biotic or abiotic measurements will be moved into the extendedMeasurementOrFact table. Before proceeding to the publishing stage, there are a number of quality control steps to complete.
Once your data has been published, you and others can access datasets through various avenues and it becomes part of OBIS’ global database!
This may seem like a daunting process at first glance, but this manual will walk you through each step, and the OBIS community is full of helpful resources. Throughout the manual you will find tutorials and tools to guide you from start to finish through the OBIS data life cycle.
2.3.0.1 Who is responsible for each phase?
Phases 1 through 3 are the responsibilities of the data provider, while Phases 3 and 4 are shared between the data provider and the node manager. Data users are involved in Phases 5 and 6.
The OBIS Secretariat is responsible for data processing and harvesting published resources.
2.4 Biodiversity data standards
From the very beginning, OBIS has championed the use of international standards for biogeographic data. Without agreement on the application of standards and protocols, OBIS would not have been able to build a large central database. OBIS uses the following standards:
The following pages of this manual review each of these in turn. We show you how to apply these standards to format your data in the Data Formatting section.
We also provide some dataset examples for your reference.
2.4.1 Darwin Core
Contents
2.4.1.1 Introduction to Darwin Core
Darwin Core is a body of standards (i.e., identifiers, labels, definitions) that facilitate sharing biodiversity informatics. It provides stable terms and vocabularies related to biological objects/data and their collection. Darwin Core is maintained by TDWG (Biodiversity Information Standards, formerly The International Working Group on Taxonomic Databases). Stable terms and vocabularies are important for ensuring the datasets in OBIS have consistently interpretable fields. By following Darwin Core standards, both data providers and users can be certain of the definition and quality of data.
2.4.1.1.1 History of Darwin Core and OBIS
The old OBIS schema was an OBIS extension to Darwin Core 1.2., which was based on Simple Darwin Core, a subset of Darwin Core which does not allow any structure beyond rows and columns. This old schema added some terms which were important for OBIS, but were not supported by Darwin Core at the time (e.g., start and end date and start and end latitude and longitude, depth range, lifestage, and terms for abundance, biomass and sample size).
In 2009, the Executive Committee of TDWG announced their ratification of an updated version of Darwin Core as a TDWG Standard. Ratified Darwin Core unifies specializations and innovations emerging from diverse communities, and provides guidelines for ongoing enhancement. The Darwin Core Quick Reference Guide links to TDWG’s term definitions and related practices for Ratified Darwin Core. We will discuss the relevance of terms in this guide further below.
In December 2013, the 3rd session of the IODE Steering Group for OBIS agreed to transition OBIS globally to the TDWG-Ratified version of Darwin Core, and the mapping of the (old) OBIS specific terms to Darwin Core can be found here.
2.4.1.2 Darwin Core (DwC) terms
DwC terms correspond to the column names of your dataset and can be grouped according to class type for convenience, e.g., Taxa, Occurrence, Record, Location, etc. It is important to use DwC field names because only columns using Darwin Core terms as headers will be recognized.
A list of all possible Darwin Core terms can be found on TDWG. However, OBIS does not parse all terms (note this doesn’t mean you cannot include them, they just will not be parsed when you publish to OBIS). Below is an overview of the most relevant Darwin Core terms to consider when contributing to OBIS, with guidelines regarding their use. We have also compiled a convenient checklist of OBIS-accepted terms, their DwC class type, and which OBIS file (Event Core, Occurrence, eMoF, etc.) it is likely to be found in.
Note that OBIS currently has seven required and one strongly recommended DwC term: occurrenceID, eventDate, decimalLongitude, decimalLatitude, scientificName, occurrenceStatus, basisOfRecord, scientificNameID (strongly recommended).
The following DwC terms are related to the Class Taxon:
- scientificName
- scientificNameID
- scientificNameAuthorship
- kingdom
- taxonRank
- taxonRemarks
The following DwC terms are related to the Class Identification:
- identifiedBy
- dateIdentified
- identificationReferences
- identificationRemarks
- identificationQualifier
- typeStatus
The following DwC terms are related to the Class Occurrence:
- occurrenceID
- occurrenceStatus
- recordedBy
- individualCount (OBIS recommends to add measurements to eMoF)
- organismQuantity (OBIS recommends to add measurements to eMoF)
- organismQuantityType (OBIS recommends to add measurements to eMoF)
- sex (OBIS recommends to add measurements to eMoF)
- lifeStage (OBIS recommends to add measurements to eMoF)
- behavior
- associatedTaxa
- occurrenceRemarks
- associatedMedia
- associatedReferences
- associatedSequences
- catalogNumber
- preparations
The following DwC terms are related to the Class Record level:
- basisOfRecord
- institutionCode
- collectionCode
- collectionID
- bibliographicCitation
- modified
- dataGeneralizations
The following DwC terms are related to the Class Location:
- decimalLatitude
- decimalLongitude
- coordinateUncertaintyInMeters
- geodeticDatum
- footprintWKT
- minimumDepthInMeters
- maximumDepthInMeters
- minimumDistanceAboveSurfaceInMeters
- maximumDistanceAboveSurfaceInMeters
- locality
- waterBody
- islandGroup
- island
- country
- locationAccordingTo
- locationRemarks
- locationID
The following DwC terms are related to the Class Event:
- parentEventID
- eventID
- eventDate
- type
- habitat
- samplingProtocol (OBIS recommends to add sampling facts to eMoF)
- sampleSizeValue (OBIS recommends to add sampling facts to eMoF)
- SampleSizeUnit (OBIS recommends to add sampling facts to eMoF)
- samplingEffort (OBIS recommends to add sampling facts to eMoF)
The following DwC terms are related to the Class MaterialSample:
- materialSampleID
2.4.1.3 Darwin Core guidelines
2.4.1.3.1 Taxonomy and identification
scientificName (required term) should always contain the originally recorded scientific name, even if the name is currently a synonym. This is necessary to be able to track back records to the original dataset. The name should be at the lowest possible taxonomic rank, preferably at species level or lower, but higher ranks, such as genus, family, order, class etc. are also acceptable. We recommend to not include authorship in scientificName, and only use scientificNameAuthorship for that purpose. The scientificName term should only contain the name and not identification qualifications (such as ?, confer or affinity), which should instead be supplied in the IdentificationQualifier term, see examples below. taxonRemarks can capture comments or notes about the taxon or name.
A WoRMS LSID should be added in scientificNameID (strongly recommended term), OBIS will use this identifier to pull the taxonomic information from the World Register of Marine Species (WoRMS) into OBIS and attach it to your dataset. This information includes:
- Taxonomic classification (kingdom through species)
- The accepted name in case of invalid names or synonyms
- AphiaID
- IUCN red list category
LSIDs are persistent, location-independent, resource identifiers for uniquely naming biologically significant resources. More information on LSIDs can be found at www.lsid.info. For example, the WoRMS LSID for Solea solea is: urn:lsid:marinespecies.org:taxname:127160, and can be found at the bottom of each WoRMS taxon page, e.g. Solea solea.
kingdom and taxonRank can help us in identifying the provided scientificName in case the name is not available in WoRMS. kingdom in particular can help us find alternative genus-species combinations and avoids linking the name to homonyms. Please contact the WoRMS data management team (info@marinespecies.org) in case the scientificName is missing in WoRMS. kingdom and taxonRank are not necessary when a correct scientificNameID is provided.
OBIS recommends providing information about how an identification was made, for example by which ID key, species guide or expert; and by which method (e.g morphology vs. genomics), etc. The person’s name who made the taxonomic identification can go in identifiedBy and when in dateIdentified. Use the ISO 8601:2004(E) standard for date and time, for instructions see Time. A list of references, such as field guides used for the identification can be listed in identificationReferences. Any other information, such as identification methods, can be added to identificationRemarks.
Examples:
| scientificNameID | scientificName | kingdom | phylum | class |
|---|---|---|---|---|
| urn:lsid:marinespecies.org:taxname:142004 | Yoldiella nana | Animalia | Mollusca | Bivalvia |
| urn:lsid:marinespecies.org:taxname:140584 | Ennucula tenuis | Animalia | Mollusca | Bivalvia |
| urn:lsid:marinespecies.org:taxname:131573 | Terebellides stroemii | Animalia | Annelida | Polychaeta |
| order | family | genus | specificEpithet | scientificNameAuthorship |
|---|---|---|---|---|
| Nuculanoida | Yoldiidae | Yoldiella | nana | (Sars M., 1865) |
| Nuculoida | Nuculidae | Ennucula | tenuis | (Montagu, 1808) |
| Terebellida | Trichobranchidae | Terebellides | stroemii | Sars, 1835 |
If the record represents a nomenclatural type specimen, the term typeStatus can be used, e.g. for holotype, syntype, etc.
In case of low confidence identifications, and the scientific name contains qualifiers such as cf., ? or aff., then this name should go in identificationQualifier, and scientificName should contain the name of the lowest possible taxon rank that refers to the most accurate identification. E.g. if the specimen was accurately identified down to genus level, but not species level, then the scientificName should contain the name of the genus, the scientificNameID should contain the LSID the genus and the identificationQualifier should contain the low confidence species name combined with ? or other qualifiers. The table below shows a few examples:
The use and definitions for additional ON signs (identificationQualifier) can be found in Open Nomenclature in the biodiversity era, which provides examples for using the main Open Nomenclature qualifiers associated with physical specimens. The publication Recommendations for the Standardisation of Open Taxonomic Nomenclature for Image-Based Identifications provides examples and definitions for identificationQualifiers for non-physical specimens (image-based).
Examples:
| scientificName | scientificNameAuthorship | scientificNameID | taxonRank | identificationQualifier | taxonConceptID |
|---|---|---|---|---|---|
| Pelagia | Péron & Lesueur, 1810 | urn:lsid:marinespecies.org:taxname:135262 | genus | gen. nov. | Pelagia gen. nov. |
| Pelagia benovici | Piraino, Aglieri, Scorrano & Boero, 2014 | urn:lsid:marinespecies.org:taxname:851656 | species | sp. nov | Pelagia benovici sp. nov |
| Gadus | Linnaeus, 1758 | urn:lsid:marinespecies.org:taxname:125732 | genus | cf. morhua | Gadus cf. morhua |
| Polycera | Cuvier, 1816 | urn:lsid:marinespecies.org:taxname:138369 | genus | cf. hedgpethi | Polycera cf. hedgpethi |
| Tubifex | Lamarck, 1816 | urn:lsid:marinespecies.org:taxname:137392 | genus | ? | Tubifex tubifex(Müller, 1774)? |
| Tubifex | Lamarck, 1816 | urn:lsid:marinespecies.org:taxname:137392 | genus | sp. inc. | Tubifex tubifex(Müller, 1774)sp. inc. |
| Brisinga | Asbjørnsen, 1856 | urn:lsid:marinespecies.org:taxname:123210 | genus | gen. inc. | Brisinga gen. inc. |
| Uroptychus compressus | Baba & Wicksten, 2019 | urn:lsid:marinespecies.org:taxname:1332465 | genus | sp. inc. | Uroptychus compressus sp. inc. |
| Eurythenes | S. I. Smith in Scudder, 1882 | urn:lsid:marinespecies.org:taxname:101607 | genus | sp. DISCOLL.PAP.JC165.674 | Eurythenes sp.DISCOLL.PAP.JC165.674 |
| Paroriza | Hérouard, 1902 | urn:lsid:marinespecies.org:taxname:123467 | genus | sp.[unique123]aff.pallens | Paroriza sp.[unique123]aff. pallens |
| Aristeidae | Wood-Mason in Wood-Mason & Alcock, 1891 | urn:lsid:marinespecies.org:taxname:106725 | family | stet. | Aristeidae stet. |
| Nematocarcinus | Milne-Edwards, 1881 | urn:lsid:marinespecies.org:taxname:107015 | genus | sp.indet. | Nematocarcinus sp.indet. |
| Brisinga | Asbjørnsen, 1856 | urn:lsid:marinespecies.org:taxname:123210 | genus | gen.inc. | Brisinga gen.inc. |
| Brisinga costata | Verrill, 1884 | urn:lsid:marinespecies.org:taxname:17825 | species | sp.inc. | Brisinga costata sp.inc. |
2.4.1.3.2 Occurrence
occurrenceID (required term) is an identifier for the occurrence record and should be persistent and globally unique. If the dataset does not yet contain (globally unique) occurrenceIDs, then they should be created. Guideline for ID creation can be found here
occurrenceStatus (required term) is a statement about the presence or absence of a taxon at a location. It is an important term, because it allows us to distinguish between presence and absence records. It is a required term and should be filled in with either present or absent.
A few terms related to quantity: organismQuantity and organismQuantityType, have been added to the TDWG ratified Darwin Core. This is a lot more versatile than the older individualCount field. However, OBIS recommends to use the Extended MeasurementorFact extension for quantitative measurements because of the standardization of terms and the fact that you can link these measurements to sampling events and factual sampling information.
Please take note that OBIS recommends all quantitative measurements and sampling facts to be placed in the ExtendedMeasurementOrFact extension and not in the Darwin Core files.
In the case specimens were collected and stored (e.g. museum collections), the catalogNumber and preparations terms can be used to provide the identifier for the record in the collection and to document the preparation and preservation methods. The term typeStatus see above (under identification) can be used in this context too.
Both associatedMedia, associatedReferences and associatedSequences are global unique identifiers or URIs pointing to respectively associated media (e.g. online image or video), associated literature (e.g. DOIs) or genetic sequence information (e.g. GenBANK ID).
associatedTaxa include a list (concatenated and separated) of identifiers or names of taxa and their associations with the Occurrence, e.g. the species occurrence was associated to the presence of kelp such as Laminaria digitata.
The recommended vocabulary for sex see BODC vocab : S10, for lifeStage see BODC vocab: S11, behavior (no vocab available), and occurrenceRemarks can hold any comments or notes about the Occurrence.
recordedBy can hold a list (concatenated and separated) of names of people, groups, or organizations responsible for recording the original Occurrence. The primary collector or observer, especially one who applies a personal identifier (recordNumber), should be listed first.
Example:
| collectionCode | occurrenceID | catalogNumber | occurrenceStatus |
|---|---|---|---|
| SluiceDock_benthic_1976/1981 | SluiceDock_benthic_1976_1 | SluiceDock_benthic_1976_1 | present |
| SluiceDock_benthic_1976/1981 | SluiceDock_benthic_1976_2 | SluiceDock_benthic_1976_2 | present |
| SluiceDock_benthic_1976/1981 | SluiceDock_benthic_1979-07/1980-06_1 | SluiceDock_benthic_1979-07/1980-06_1 | present |
2.4.1.3.3 Record level terms
basisOfRecord (required term) specifies the nature of the record, i.e. whether the occurrence record is based on a stored specimen or an observation. In case the specimen is collected and stored in a collection (e.g. at a museum, university, research institute), the options are:
PreservedSpecimene.g. preserved in ethanol, tissue etc.FossilSpecimena fossil, which allows OBIS to make the distinction between the date of collection and the time period the specimen was assumed aliveLivingSpecimenan intentionally kept/cultivated living specimen e.g. in an aquarium or culture collection.
In case no specimen is deposited, the basis of record is either HumanObservation (e.g bird sighting, benthic sample but specimens were discarded after counting), or MachineObservation (e.g. for occurrences based on automated sensors such as image recognition, etc). For records pertaining to genetic samples, basisOfRecord can be MaterialSample (e.g. in the DNA-derived data extension).
When the basisOfRecord is either a preservedSpecimen, LivingSpecimen or FossilSpecimen please also add the institutionCode, collectionCode and catalogNumber, which will enable people to visit the collection and re-examine the material. Sometimes, for example in case of living specimens, a dataset can contain records pointing to the origin, the in-situ sampling position as well as a record referring to the ex-situ collection. In this case please add the event type information in eventRemarks (see OBIS manual: event).
institutionCode identifies the custodian institute (often by acronym), collectionCode identifies the collection or dataset within that institute. Collections cannot belong to multiple institutes, so all records within a collection should have the same institutionCode. The collectionID is an identifier for the record within the dataset or collection.
bibliographicCitation allows for providing different citations on record level, while a single citation for the entire dataset can and should be provided in the metadata (see EML). The citation at record level can have the format of a chapter in a book, where the book is the dataset citation. The record citation will have preference over the dataset citation. We do not, however, recommend to create different citations for every record, as this will explode the number of citations and will hamper the re-use of data.
modified is the most recent date-time on which the resource was changed. It is required to use the ISO 8601:2004(E) standard, for instructions see Time.
dataGeneralizations refers to actions taken to make the shared data less specific or complete than in its original form. Suggests that alternative data of higher quality may be available on request. This can be the case for occurrences of vulnerable or endangered species and there positions are converted to the center of grid cells.
2.4.1.3.4 Location
decimalLatitude and decimalLongitude (required terms) are the geographic latitude and longitude (in decimal degrees), using the spatial reference system given in geodeticDatum of the geographic center of a Location. The number of decimals should be appropriate for the level of uncertainty in coordinateUncertaintyInMeters (at least within an order of magnitude). coordinateUncertaintyInMeters is the radius of the smallest circle around the given position containing the whole location. Regarding decimalLatitude, positive values are north of the Equator, negative values are south of it. All values lie between -90 and 90, inclusive. Regarding decimalLongitude, positive values are east of the Greenwich Meridian, negative values are west of it. All values lie between -180 and 180, inclusive.
In OBIS, the spatial reference system to be documented in geodeticDatum is EPSG:4326. Coordinates in degrees/minutes/seconds can be converted to decimal degrees using our coordinates tool. We also provide a tool to check coordinates or to determine coordinates for a location (point, transect or polygon) on a map. This tool also allows geocoding location names using marineregions.org.
The name of the place or location can be provided in locality, and if possible linked by a locationID using a persistent ID from a gazetter, such as the MRGID from MarineRegions. If the species occurrence only contains the name of the locality, but not the exact coordinates, we recommend using a geocoding service to obtain the coordinates. Marine Regions has a search interface for geographic names, and provides coordinates and often precision in meters, which can go into coordinateUncertaintyInMeters. Another option is to use the Getty Thesaurus of Geographic Names or Google Maps: after looking up a location, the decimal coordinates can be found in the page URL. Additional information about the locality can also be stored in DwC terms such as waterBody, islandGroup, island and country. locationAccordingTo should provide the name of the gazetteer that is used to obtain the coordinates for the locality.
locationID is an identifier for the set of location information (e.g. station ID, or MRGID from marineregions), for example the Balearic Plain has MRGID: http://marineregions.org/mrgid/3956.
A Well-Known Text (WKT) representation of the shape of the location can be provided in footprintWKT. This is particularly useful for tracks, transects, tows, trawls, habitat extent or when an exact location is not known. WKT strings can be created using our WKT tool. This tool also calculates a midpoint and a radius, which can then be added to decimalLongitude, decimalLatitude, and coordinateUncertaintyInMeters respectively. There is also an R tool to calculate the centroid and radius for WKT polygons. wktmap.com can be used to visualize and share WKT strings.
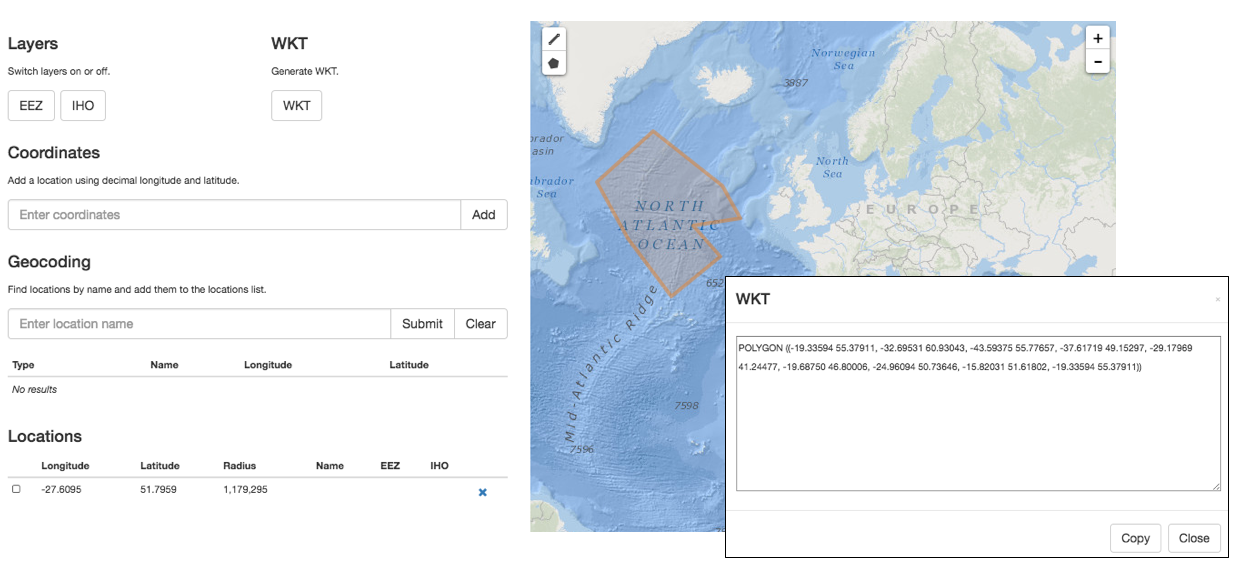
Some examples of WKT strings:
LINESTRING (30 10, 10 30, 40 40)
POLYGON ((30 10, 40 40, 20 40, 10 20, 30 10))
MULTILINESTRING ((10 10, 20 20, 10 40),(40 40, 30 30, 40 20, 30 10))
MULTIPOLYGON (((30 20, 45 40, 10 40, 30 20)),((15 5, 40 10, 10 20, 5 10, 15 5)))Example:
| decimalLatitude | decimalLongitude | geodeticDatum | coordinateUncertaintyInMeters | footprintWKT | footprintSRS |
|---|---|---|---|---|---|
| 38.698 | 20.95 | EPSG:4326 | 75033.17 | LINESTRING (20.31 39.15, 21.58 38.24) | EPSG:4326 |
| 42.72 | 15.228 | EPSG:4326 | 154338.87 | LINESTRING (16.64 41.80, 13.82 43.64) | EPSG:4326 |
| 39.292 | 20.364 | EPSG:4326 | 162083.27 | LINESTRING (19.05 40.34, 21.68 38.25) | EPSG:4326 |
Keep in mind while filling in minimumDepthInMeters and maximumDepthInMeters that this should be the depth at which the sample was taken and not the water column depth at that location. When fillling in any depth fields (minimumDepthInMeters, maximumDepthInMeters, minimumDistanceAboveSurfaceInMeters, and maximumDistanceAboveSurfaceInMeters), you should also consider which information is needed to fully understand the data. In most cases (e.g. scenario 1 and 4 in the figure below), providing minimumDepthInMeters and maximumDepthInMeters is sufficient for observations of organisms at particular depths. However, in cases where an occurrence is above the sea surface, e.g. flying birds (scenario 2 and 5), you should populate minimumDistanceAboveSurfaceInMeters, maximumDistanceAboveSurfaceInMeters, and, where relevant, you should also include minimumElevationInMeters and maximumElevationInMeters.
The minimumDistanceAboveSurfaceInMeters and maximumDistanceAboveSurfaceInMeters is the distance, in meters, above or below a reference surface or reference point. The reference surface is determined by the depth or elevation. If the depth and elevation are 0, then the reference surface is the sea surface. If a depth is given, the reference surface is the location of the depth. This can be especially useful for sediment cores taken from the sea bottom (scenario 3 in figure below). If no depth is given, then the elevation is the reference surface (scenario 5).
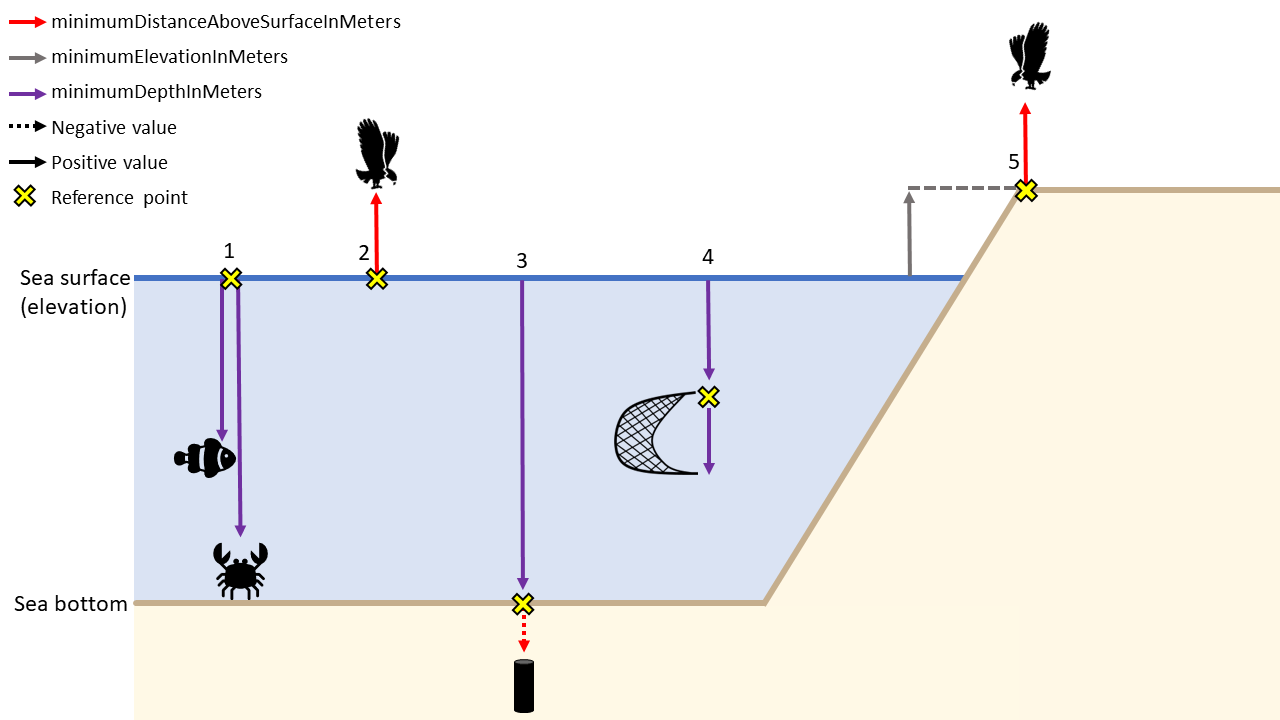
Depth scenario examples:
| Scenario | minimumDepthInMeters | maximumDepthInMeters | minimumDistanceAboveSurfaceInMeters | maximumDistanceAboveSurfaceInMeters | minimumElevationInMeters | maximumElevationInMeters |
|---|---|---|---|---|---|---|
| 1 | 40, 90 | 50, 100 | - | - | 0 | 0 |
| 2 | 0 | 0 | 10 | 15 | 0 | 0 |
| 3 | 100 | 100 | 0 | -1.5 | 0 | 0 |
| 4 | 20 | 22 | - | - | 0 | 0 |
| 5 | 0 | 0 | 10 | 15 | 10 | 10 |
2.4.1.3.5 Event
eventID is an identifier for the sampling or observation event. parentEventID is an identifier for a parent event, which is composed of one or more sub-sampling (child) events (eventIDs). See identifiers for details on how these terms can be constructed.
habitat is a category or description of the habitat in which the Event occurred (e.g. benthos, seamount, hydrothermal vent, seagrass, rocky shore, intertidal, ship wreck etc.)
2.4.1.3.6 Time
The date and time at which an occurrence was recorded goes in eventDate. This term uses the ISO 8601 standard and OBIS recommends using the extended ISO 8601 format with hyphens.

More specific guidelines on formatting dates and times can be found in the Common Data formatting issues page
2.4.1.3.7 Sampling
Information on sampleSizeValue and sampleSizeUnit is very important when an organism quantity is specified. However, with OBIS-ENV-DATA it was felt that the extended MeasurementorFact (eMoF) extension would be better suited than the DwC Event Core to store the sampled area and/or volume because in some cases sampleSize by itself may not be detailed enough to allow interpretation of the sample. For instance, in the case of a plankton tow, the volume of water that passed through the net is relevant. In case of Niskin bottles, the volume of sieved water is more relevant than the actual volume in the bottle. In these examples, as well as generally when recording sampling effort for all protocols, eMoF enables greater flexibility to define parameters, as well as the ability to describe the entire sample and treatment protocol through multiple parameters. eMoF also allows you to standardize your terms to a controlled vocabulary.
The next chapter deals with the metadata (description of the dataset) in Ecological Metadata Language.
2.4.2 Darwin Core Archive
Contents
- Darwin Core Archive
- OBIS holds more than just species occurrences: the ENV-DATA approach
- Recommended reading
2.4.2.1 Darwin Core Archive
Darwin Core Archive (DwC-A) is the standard for packaging and publishing biodiversity data using Darwin Core terms. It is the preferred format for publishing data in OBIS and GBIF. The format is described in the Darwin Core text guide. A Darwin Core Archive contains a number of text files, including data tables formatted as CSV.
The conceptual data model of the Darwin Core Archive is a star schema with a single core table, for example containing occurrence records or event records, at the center of the star. Extension tables can optionally be associated with the core table. It is not possible to link extension tables to other extension tables (to form a so-called snowflake schema). There is a one-to-many relationship between the core and extension records, so each core record can have zero or more extension records linked to it, and each extension record must be linked to exactly one core record. Definitions for the core and extension tables can be found here.
Besides data tables, a Darwin Core Archive also contains two XML files: one file which describes the archive and data file structure (meta.xml), and one file which contains the dataset’s metadata (eml.xml).
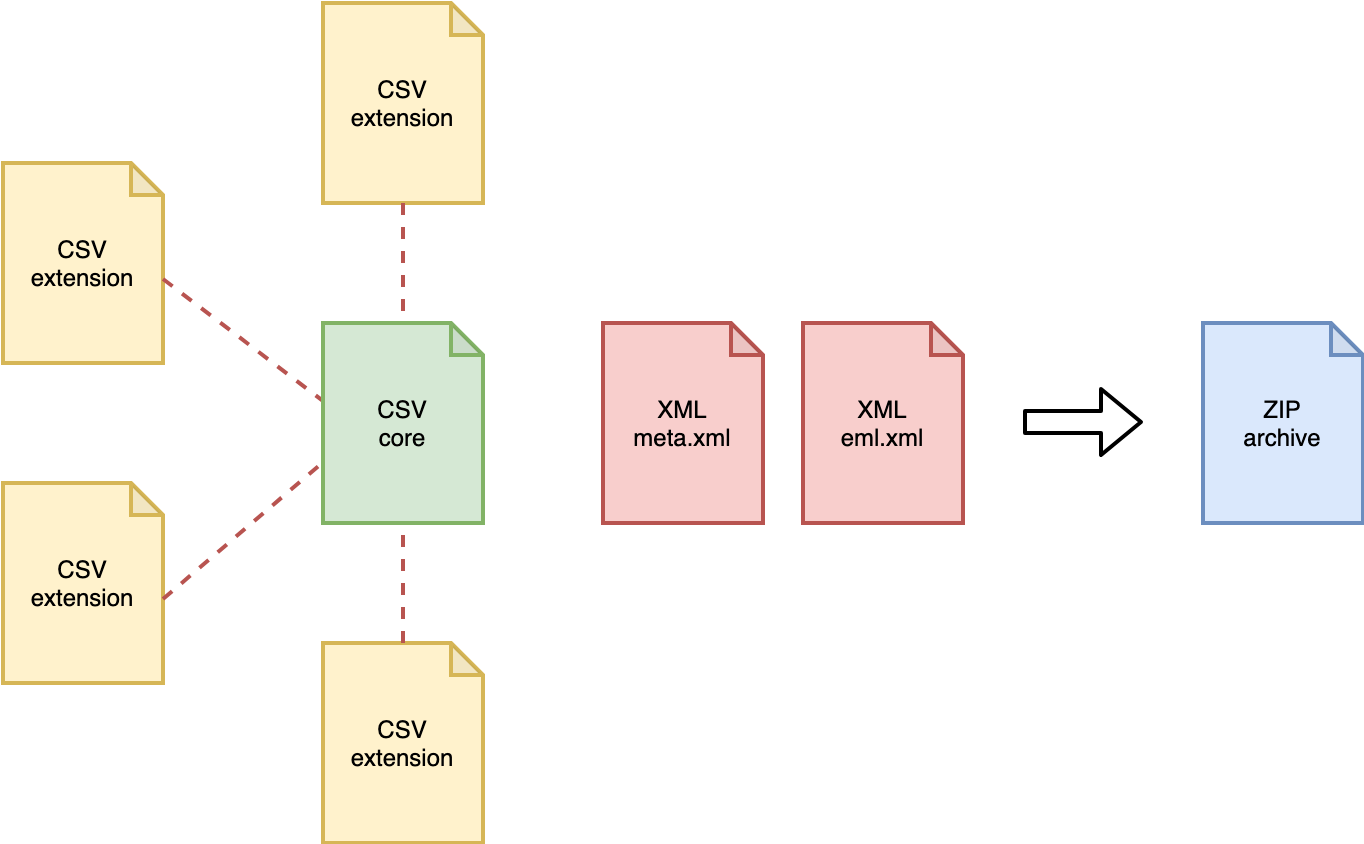
Figure: structure of a Darwin Core Archive.
2.4.2.2 OBIS holds more than just species occurrences: the ENV-DATA approach
Data collected as part of marine biological research often include measurements of habitat features (such as physical and chemical parameters of the environment), biotic and biometric measurements (such as body size, abundance, biomass), as wel as details regarding the nature of the sampling or observation methods, equipment, and sampling effort.
In the past, OBIS relied solely on the Occurrence Core, and additional measurements were added in a structured format (e.g. JSON) in the Darwin Core term dynamicProperties inside the occurrence records. This approach had significant downsides: the format is difficult to construct and deconstruct, there is no standardization of terms, and attributes which are shared by multiple records (think sampling methodology) have to be repeated many times. The formatting problem can be addressed by moving measurements to a MeasurementOrFacts extension table, but that doesn’t solve the redundancy and standardization problems.
With the release and adoption of a new core type Event Core it became possible to associate measurements with nested events (such as cruises, stations, and samples), but the restrictive star schema of Darwin Core archive prohibited associating measurements with the event records in the Event core as well as with the occurrence records in the Occurrence extension. For this reason an extended version of the existing MeasurementOrFact extension was created.
2.4.2.2.1 ExtendedMeasurementOrFact Extension (eMoF)
As part of the IODE pilot project Expanding OBIS with environmental data OBIS-ENV-DATA, OBIS introduced a custom ExtendedMeasurementOrFact or eMoF extension, which extends the existing MeasurementOrFact extension with 4 new terms:
occurrenceIDmeasurementTypeIDmeasurementValueIDmeasurementUnitID
The occurrenceID term is used to circumvent the limitations of the star schema, and link measurement records in the ExtendedMeasurementOrFact extension to occurrence records in the Occurrence extension. Note that in order to comply with the Darwin Core Archive standard, these records still need to link to an event record in the Event core table as well. Thanks to this term we can now store a variety of measurements and facts linked to either events or occurrences:
- organism quantifications (e.g. counts, abundance, biomass, % live cover, etc.)
- species biometrics (e.g. body length, weight, etc.)
- facts documenting a specimen (e.g. living/dead, behaviour, invasiveness, etc.)
- abiotic measurements (e.g. temperature, salinity, oxygen, sediment grain size, habitat features)
- facts documenting the sampling activity (e.g. sampling device, sampled area, sampled volume, sieve mesh size).
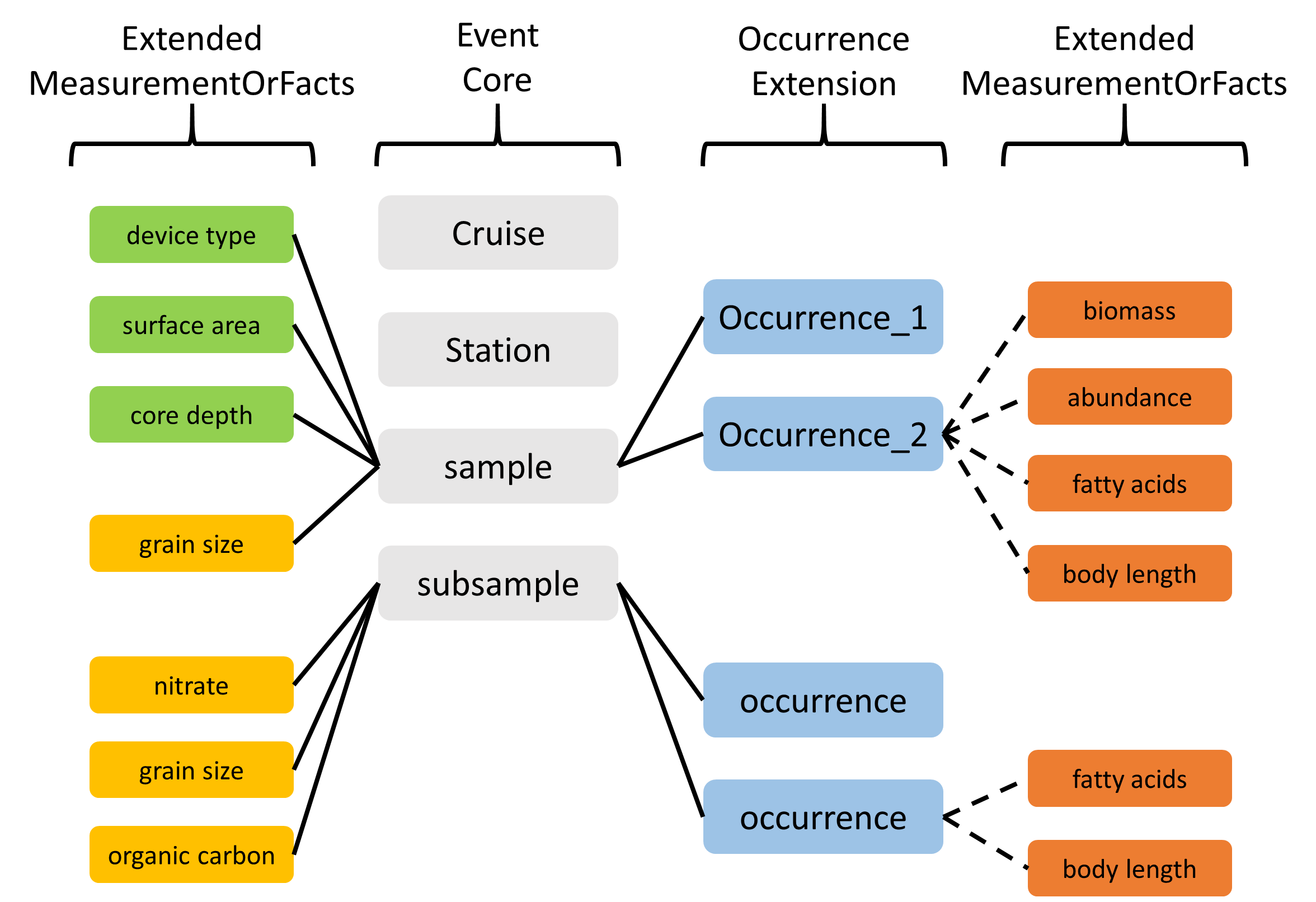
Figure: Overview of an OBIS-ENV-DATA format. Sampling parameters, abiotic measurements, and occurrences are linked to events using the eventID (full lines). Biotic measurements are linked to occurrences using the new occurrenceID field of the ExtendedMeasurementOrFact Extension (dashed lines).
2.4.2.2.2 eDNA & DNA derived data Extension
DNA derived data are increasingly being used to document taxon occurrences. To ensure these data are useful to the broadest possible community, GBIF published a guide entitled Publishing DNA-derived data through biodiversity data platforms. This guide is supported by the DNA derived data extension for Darwin Core, which incorporates MIxS terms into the Darwin Core standard. eDNA and DNA derived data is linked to occurrence data with the use of occurrenceID and/ or eventID. Refer to the Examples: ENV-DATA and DNA derived data for use case examples of eDNA and DNA derived data.
2.4.2.2.3 A special case: habitat types
Including information on habitats (biological community, biotope, or habitat type) is possible and encouraged with the use of Event Core. However, beware the unconstrained nature of the terms measurementTypeID, measurementValueID, and measurementUnitID which can lead to inconsistently documented habitat measurements within the Darwin Core Archive standard. To ensure this data is more easily discoverable, understood or usable, refer to Examples: habitat data and/or Duncan et al. (2021) for use case examples and more details.
2.4.2.2.4 Recommended reading
- De Pooter et al. 2017. Toward a new data standard for combined marine biological and environmental datasets - expanding OBIS beyond species occurrences. Biodiversity Data Journal 5: e10989. hdl.handle.net/10.3897/BDJ.5.e10989
- Duncan et al. (2021). A standard approach to structuring classified habitat data using the Darwin Core Extended Measurement or Fact Extension. EMODnet report. (Note you must refine search to Technical Reports from 2021 to identify Duncan et al.’s report)
2.4.3 Relational databases: the underlying framework of OBIS
If you are not familiar with relational databases, it can be difficult to understand the underlying framework OBIS relies on. This section will help you understand relational databases, how they relate to OBIS, the data you will format for OBIS, and the data you may download from OBIS.
Why do we use relational databases in the first place? You are probably familiar with flat databases which contain all data in one table - this is likely how your own data are formatted. Relational databases instead consist of multiple data tables that each contain related information. When all this information is presented in one table, the table becomes larger, very complicated, and the likelihood of data duplication increases. Relational databases seek to simplify complexities and reduce redundancy by allowing information to be self-contained, but linked to each other.
You can think of a relational database as separate Excel sheets or data tables that are related to each other. One data table could be a “core” table, whereas others are “extensions”. Sometimes the relationships between core and extension tables are hierarchical, but this is not always the case. There is, however, always a relationship linking core and extension tables.
Let’s review core and extension tables and how we use them for OBIS.
Core tables contain information that is applicable to all extension tables, and extension tables contain more information about the records within the Core table. Each table, whether core or extension, contains records and attributes. Each row is a record (e.g., a sampling event, a species’ occurrence), whereas each column is an attribute (e.g., a date, a measurement).
Records between tables are linked to each other by the use of identifiers. A description of measurements pertaining to a record in an Extension table will have the same identifier as the record it is describing in the Core table. By using identifiers to link records, we reduce data repetition, see below for examples. In the Darwin Core format that OBIS uses, the core table is either Event or Occurrence, and datasets can have one, none, or more extension tables. Further explanation of data formatting in OBIS is covered in the Data Formatting section of the OBIS manual.
Let’s review an example to fully understand how relational databases work. We will look at a simple relational database used by a fictional country that tracks student performance in three different courses between three schools. Rather than trying to contain information about each school, course, and student performance in one place, this information is split into three separate tables. We see that the pink table gives us information about each school - its name, and the district it belongs to. Each school also has a schoolID, an identifier linking to the blue table where we can see student performance (course mean) in each course, the class size, and year. You will notice that the course mean and class size are bundled under columns called measurementType and measurementValue. These are similar to the eMoF vocabularies and are integral to reducing repeated data, especially when one dataset has reoccurring information. Finally we see that the courseID in the blue table links to the yellow one with the courseID identifier, giving us information about each course.
A fourth table could easily be created to track total school population size through time. In contrast, if this information was only presented in the pink Schools in Country table, the school information would be duplicated as you add rows for each year. In this way, you can easily see how useful relational databases are. Of course, this is a simplified example but it demonstrates how related tables can be linked by identifiers to reduce table complexity and data replication.
We elaborate on how this structure is applied within OBIS here.
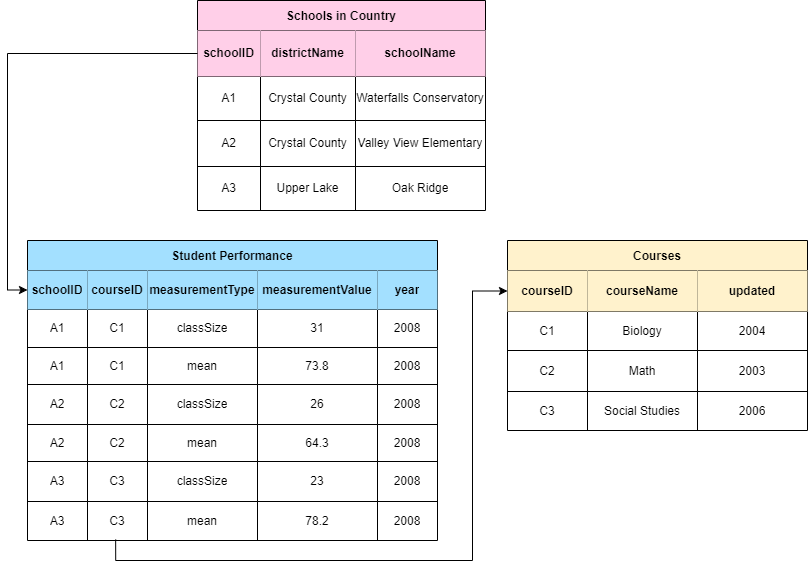
Note that when OBIS harvests data, datasets are flattened - i.e., all separate data tables are combined into one. This is the kind of file you will receive when you download data from OBIS. The reason for this is that querying relational databases significantly reduces computational time, as opposed to querying a flat database. Relational databases also facilitate requests for subsets that meet particular criteria - e.g., all data from Norway for one species above a certain depth.
2.4.3.1 How to avoid redundancy
Avoiding redundancy and data duplication within your dataset is built into the OBIS data structure. Utilizing the ENV-DATA approach, which delineates relationships between the core table and extension tables, we can limit the repetition of data.
For example, let us consider the dates of a ship cruise where a series of bottom trawls were taken. The sampling information (e.g., date range, equipment used, etc.) for each species collected in these trawls is the same. Because of this, we know we are dealing with unique sampling events and thus we will use Event core. So, our Event core table will contain all information related to the sampling events (e.g., date, location). Then, information pertaining to each collected species (e.g., abundance, biomass, sampling methods, etc.) will be placed in an extension, the (Extended)MeasurementOrFact table. Here, each measurement for each species and sample will occur on a separate record. These records will be linked to the correct sampling event in the Event core by an identifier - the eventID. If we were to put this data in one file, the fields related to date and location (e.g., eventDate, decimalLongitude, decimalLatitude, etc.) would be repeated for each species.
Let’s consider another example. If you took one temperature measurement from the water column where you took your sample, each species found in that sample would have the same temperature measurement. By linking such measurements to the event instead of each occurrence, we are able to reduce the amount of data being repeated.
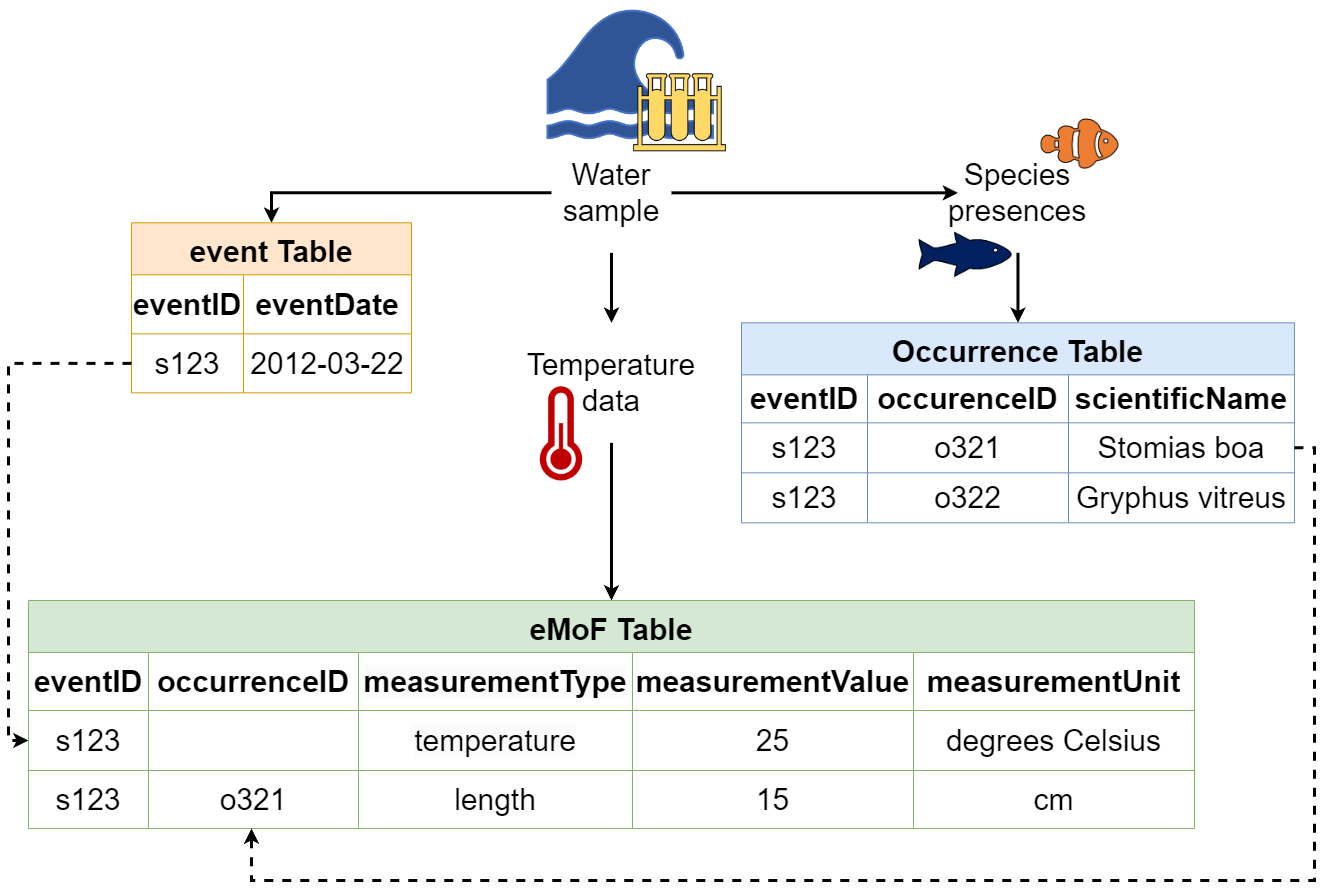
An advantage of structuring data this way is that if any mistakes are made, you only need to correct it once! So you can see that using relational event structures (when applicable) in combination with extension files can really simplify and reduce the number of times data are repeated.
Caveat: However we would like to note that in some cases, data duplication may occur due to the star schema structure. For example, when publishing DNA-derived data, Occurrence core will have to be used, which necessitates the repetition of event data for each occurrence record.
2.4.4 Ecological Metadata Language
OBIS (and GBIF) uses the Ecological Metadata Language (EML) as its metadata standard, which is specifically developed for the earth, environmental and ecological sciences. It is based on prior work done by the Ecological Society of America and associated efforts. EML is implemented as XML. See more information on EML.
OBIS uses the GBIF EML profile (version 1.1). In case data providers use ISO19115/ISO19139, there is a mapping available here.
For OBIS, the following 4 terms are the bare minimum required: Title, Citation, Contact and Abstract. Below is an overview of all the EML terms used to describe datasets:
title [xml:lang="..."]: A good descriptivetitleis indispensable and can provide the user with valuable information, making the discovery of data easier. Multiple titles may be provided, particularly when trying to express the title in more than one language (use the “xml:lang” attribute to indicate the language if not English/en).creator;metadataProvider;associatedParty;contact: These are the people and organizations responsible for the dataset resource, either as the creator, the metadata provider, contact person or any other association. The following details can be provided:individualNamegivenNamesurName
organizationName: Name of the institution.positionName: to be used as alternative to persons names (leaveindividualNameblank and usepositionNameinstead e.g. data manager).addressdeliveryPointcityadministrativeAreapostalCodecountry
phone
electronicMailAddressonlineUrl: personal websiterole: used withassociatedPartyto indicate the role of the associated person or organization.userID: e.g. ORCID.directory
pubDate: The date that the resource was published. Use ISO 8601.language: The language in which the resource (not the metadata document) is written. Use ISO language code.abstract: Brief description of the data resource.para
keywordSetkeyword: Note only one keyword per keyword field is allowed.keywordThesaurus: e.g. ASFA
additionalInfo: OBIS checks this EML field for harvesting. It should contain marine, harvested by iOBIS.para
coveragegeographicCoveragegeographicDescription: a short text description of the area. E.g. the river mounth of the Scheldt Estuary.boundingCoordinateswestBoundingCoordinateeastBoundingCoordinatenorthBoundingCoordinatesouthBoundingCoordinate
temporalCoverage: Use ISO 8601singleDateTimerangeOfDatesbeginDatecalendarDate
endDatecalendarDate
taxonomicCoverage: taxonomic information about the dataset. It can include a species list.generalTaxonomicCoveragetaxonomicClassificationtaxonRankNametaxonRankValuecommonName
intellectualRights: Statement about IPR, Copyright or various Property Rights. Also read the guidelines on the sharing and use of data in OBIS.para
purpose: A description of the purpose of this dataset.para
methodsmethodStep: Descriptions of procedures, relevant literature, software, instrumentation, source data and any quality control measures taken.sampling: Description of sampling procedures including the geographic, temporal and taxonomic coverage of the study.studyExtent: Description of the specific sampling area, the sampling frequency (temporal boundaries, frequency of occurrence), and groups of living organisms sampled (taxonomic coverage).samplingDescription: Description of sampling procedures, similar to the one found in the methods section of a journal article.para
qualityControl: Description of actions taken to either control or assess the quality of data resulting from the associated method step.
projecttitleidentifierpersonnel: The personnel field is used to document people involved in a research project by providing contact information and their role in the project.descriptionfunding: The funding field is used to provide information about funding sources for the project such as: grant and contract numbers; names and addresses of funding sources.para
studyAreaDescriptiondesignDescription: The description of research design.
maintenancedescriptionpara
maintenanceUpdateFrequency
additionalMetadatametadatadateStamp: The dateTime the metadata document was created or modified (ISO 8601).metadataLanguage: The language in which the metadata document (as opposed to the resource being described by the metadata) is writtenhierarchyLevelcitation: A single citation for use when citing the dataset. The IPT can also auto-generate a citation based on the metadata (people, title, organization, onlineURL, DOI etc).bibliography: A list of citations that form a bibliography on literature related / used in the datasetresourceLogoUrl: URL of the logo associated with a dataset.parentCollectionIdentifiercollectionIdentifierformationPeriod: Text description of the time period during which the collection was assembled. E.g., “Victorian”, or “1922 - 1932”, or “c. 1750”.livingTimePeriod: Time period during which biological material was alive (for palaeontological collections).specimenPreservationMethodphysicalobjectNamecharacterEncodingdataFormatexternallyDefinedFormatformatName
distribution: URL linksonlineurl function="download"url function="information"
alternateIdentifier: It is a Universally Unique Identifier (UUID) for the EML document and not for the dataset. This term is optional.
2.4.4.1 Metadata Sections
There are several categories/pages for metadata you must provide, which includes basic information about the:
- Dataset and data provider
- Geographic/taxonomic/temporal coverage
- Keywords
- Hosting institution information
- Information regarding associated project(s)
- Sampling methods
- How to cite the dataset
- Museum collection (if applicable)
- Other external links (e.g. a homepage) or additional metadata
We review each of these sections below.
2.4.4.1.1 Title
The IPT requires you to provide a Shortname. Shortnames serve as an identifier for the resource within the IPT installation (so should be unique within your IPT), and will be used as a parameter in the URL to access the resource via the Internet. Please use only alphanumeric characters, hyphens, or underscores. E.g. largenet_im in http://ipt.vliz.be/eurobis/resource?r=largenet_im. After creating a new dataset resource, the field title will be filled out with the short name you provided earlier. Please make sure you provide a dataset title following the guidelines below.
Dataset titles provided to OBIS node managers are often very cryptic, such as an acronym, and often only understandable by the data provider. However, to increase the discoverability and be useful for a larger audience, the dataset title should be as descriptive and complete as possible. OBIS recommends titles to contain information about the taxonomic, geographic and temporal coverage. If the dataset title does not meet these criteria and you believe the title should be changed, then contact the data provider with a suggestion or ask for a more descriptive title. If the dataset has already been published (made publicly available) - and therefore known by that title elsewhere, then the same title should be kept (even if it would not meet the proposed guidelines)! Changing the title of an already published dataset cannot be done, as this will generate confusion and possible duplicates in systems like OBIS or GBIF in a later stage.
The acronym or working title could still be documented in the metadata, so there is no confusion about how the full title is linked to the originally provided acronym or working title.
Caution: Always consult the data provider when changing a dataset title to a more workable and descriptive version.
| Originally received title | Title Recommended by Node Manager |
|---|---|
| BIOCEAN | BIOCEAN database on deep sea benthic fauna |
| Biomôr | Benthic data from the Southern Irish Sea from 1989-1991 |
| Kyklades | Zoobenthos of the Kyklades (Aegean Sea) |
| REPHY | Réseau de Surveillance phytoplanctonique |
2.4.4.1.2 Abstract
The abstract or description of a dataset provides basic information on the content of the dataset. The information in the abstract should improve understanding and interpretation of the data. It is recommended that the description indicates whether the dataset is a subset of a larger dataset and – if so – provide a link to the parent metadata and/or dataset.
If the data provider or OBIS node require bi- or multilingual entries for the description (e.g. due to national obligations) then the following procedure can be followed:
- Indicate English as metadata language
- Enter the English description first
- Type a slash (/)
- Enter the description in the second language
Example: The Louis-Marie herbarium grants a priority to the Arctic-alpine, subarctic and boreal species from the province of Quebec and the northern hemisphere. This dataset is mainly populated with specimens from the province of Quebec. / L’Herbier Louis-Marie accorde une priorité aux espèces arctiques-alpines, subarctiques et boréales du Québec, du Canada et de l’hémisphère nord. Ce jeu présente principalement des spécimens provenant du Québec.
2.4.4.1.3 People and Organizations
The EML has several possible roles/functions to describe a contact, creator, metadata provider and associated party.
The contact is the person or organization that curates the resource and who should be contacted to get more information or to whom questions with the resource or data should be addressed. Although a number of fields are not required, we strongly recommend providing as much information as possible, and in particular the email address. This will also be the contact information that appears on the OBIS metadata pages.
The creator is the person or organization responsible for the original creation of the resource content. When there are multiple creators, the one that bears the greatest responsibility is the resource creator, and other people can be added as associated parties with a role such as ‘originator’, ‘content provider’, ‘principal investigator’, etc.
Possible functions/roles:
- Originator (person/organization that originally gathered/prepared the dataset)
- Content provider (principal person/organization that contributed content to the dataset)
If the resource contact and the resource creator are identical, the IPT allows you to easily copy the information.
The metadata provider is the person or organization responsible for producing the resource metadata. If the metadata are provided by the original data provider, then his/her contact details should be filled in. If no metadata are available (e.g. for historical datasets, with no contact person), then the metadata can be completed by e.g. the OBIS node manager and the OBIS node manager becomes the metadata provider.
The Associated Parties contains information about one or more people or organizations associated with the resource in addition to those already covered on the IPT Basic Metadata page. For example, if there would be multiple contact persons or metadata creators, they can be added in this IPT section. The principal contact/creator should, however, be added in the IPT Basic Metadata section, not the Associated Parties section. It is recommended to complete this section together with the IPT Basic Metadata page, to avoid confusion or overlap in added information.
Possible functions/roles for associated parties are:
- Custodian steward (person/organization responsible for/takes care of the dataset paper)
- Owner (person/organization that owns the data – may or may not be the custodian)
- Point of contact (person/organization to contact for further information on the dataset)
- principal investigator (primary scientific contact associated with the dataset)
Notes:
The owner of a dataset will, in most cases, be an institute, and not an individual person. Although the fields ‘last name’, and ‘position’ are indicated as mandatory fields, it is possible to just add the institute name in the ‘last name’ field for the role ‘owner’.
The contact persons in the metadata (contact, creator, metadata creator) are used in the dataset citation (auto-generation) and those added as ‘associated parties’ are not included as “co-authors”.
2.4.4.1.4 License and IP Rights
OBIS has published its guidelines on the sharing and use of data here. The recommended licenses for datasets published in OBIS are the Creative Commons Licenses (CC-0, CC-BY, CC-BY-NC), of which CC-0 is the most preferred and CC-BY-NC is least preferred. A Creative Commons license means:
- You are free:
- to share => to copy, distribute and use the database
- to create => to produce works from the database
- to adapt => to modify, transform and build upon the database
In case of CC-0: public domain: CC-0 is the preferred option identified by the OBIS steering group. You waive any copyright you might have over the data(set) and dedicate it to the public domain. You cannot be held liable for any (mis)use of the data either. Although CC-0 doesn’t legally require users of the data to cite the source, it does not take away the moral responsibility to give attribution, as is common in scientific research. A good blog on why using CC-0 can be found here.
In case of CC-BY: Attribution: You must attribute any public use of the database, or works produced from the database, in the manner specified in the license. For any use or redistribution of the database, or works produced from it, you must make clear to others the license of the database and keep intact any notices on the original database.
In case of CC-BY-NC: non-commercial: like CC-BY but commercial use is not allowed. This licence can be problematic when the data is re-used in scientific journals.
2.4.4.1.5 Coverage
2.4.4.1.5.1 Geographic Coverage
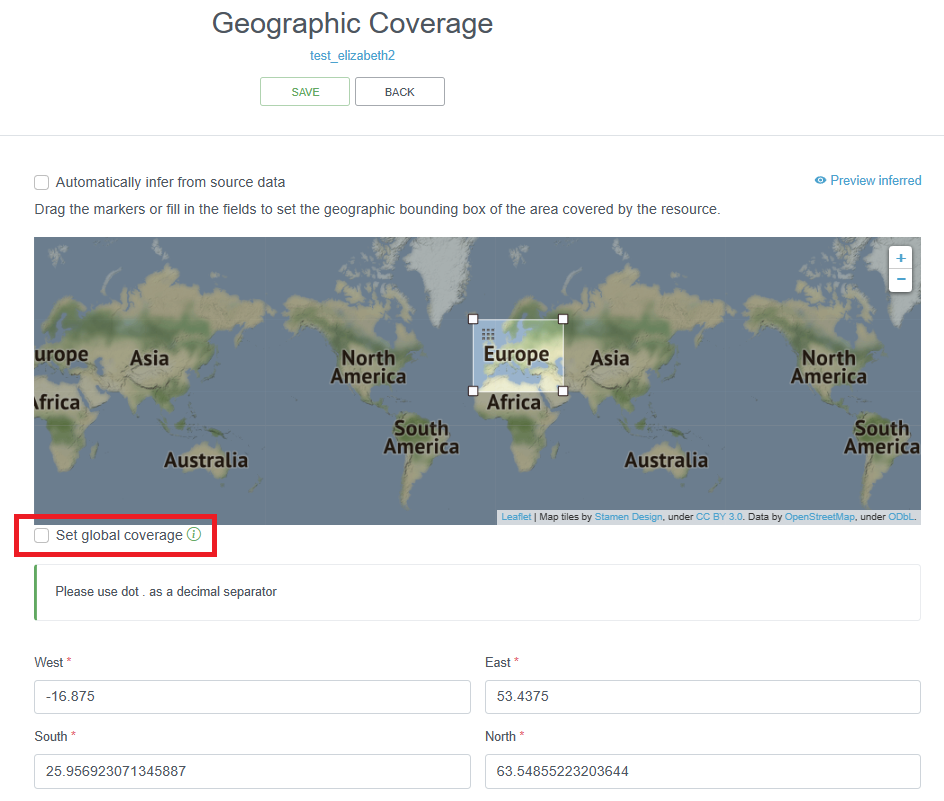
The IPT allows you to enter the geographic coverage by dragging the markers on the given map or by filling in the coordinates of the bounding box. In the description field, a more elaborate text can be provided to describe the spatial coverage indicating the larger geographical area where the samples were collected. For the latter, the sampling locations can be plotted on a map and – by making use of a Gazetteer – the wider geographical area can be derived: e.g. the relevant Exclusive Economic Zone (EEZ), IHO, FAO fishing area, Large Marine Ecosystem (LME), Marine Ecoregions of the World (MEOW), etc. The Marine Regions’ Gazetteer might prove to be a useful online tool to define the most relevant sea area(s). There are also LifeWatch Geographical Services that translate geographical positions to these wider geographical areas.
The information given in this section can also help the OBIS node manager in geographic quality control. If the geographic coverage in the EML e.g. is “North Sea”, but a number of data points are outside of this scope, then this may indicate errors, and should be checked with the data provider.
If the dataset covers multiple areas (e.g. samples from the North Sea and the Mediterranean Sea), then this should clearly be mentioned in the geographicDescription field. Note that the IPT only allows one bounding box, and you have to uncheck the “Set global coverage” box to change box bounds.
2.4.4.1.5.2 Taxonomic Coverage
This section can capture two things:
- A description of the range of taxa that are addressed in the data set. OBIS recommends to only add the higher classification (Kingdom, Class or Order) of the involved groups (e.g. Bivalvia, Cetacea, Aves, Ophiuroidea…). You can easily draw a list of higher taxonomic ranks from the WoRMS taxon match service (or ask the data provider). The taxonomic coverage is not a mandatory field, but the information stored here can be very useful as background information. The description can also contain common names, such as e.g. benthic foraminifera or mussels.
- An overview of all the involved taxa (not recommended, as all the taxa are already listed in the dataset).
Note: OBIS also recommends to add information on the (higher) taxonomic groups in the (descriptive) dataset title and abstract.
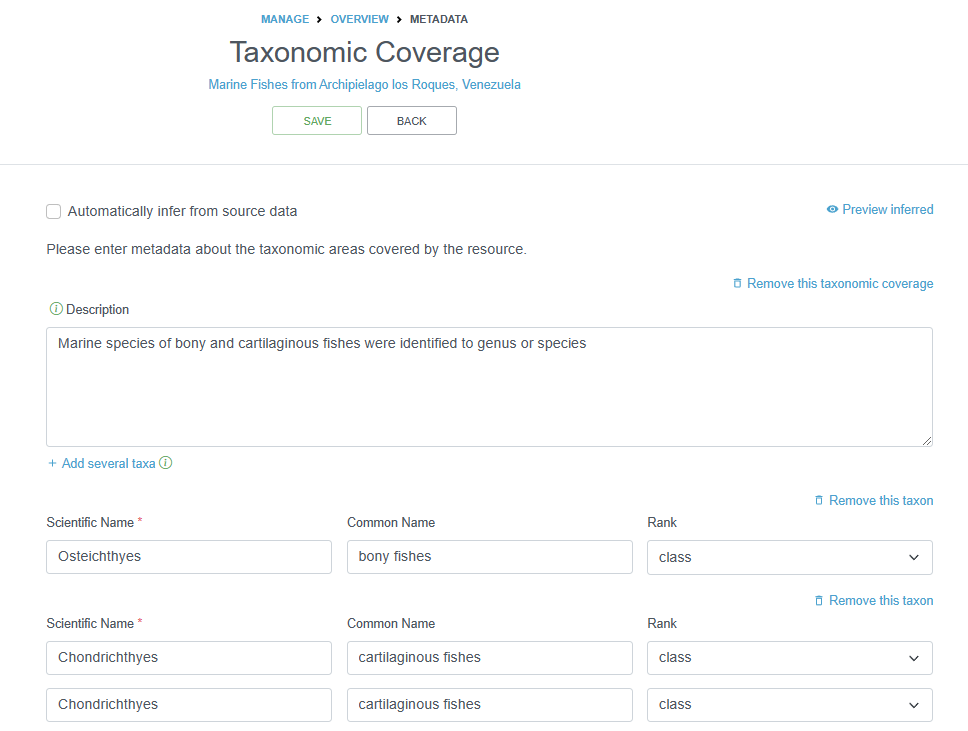
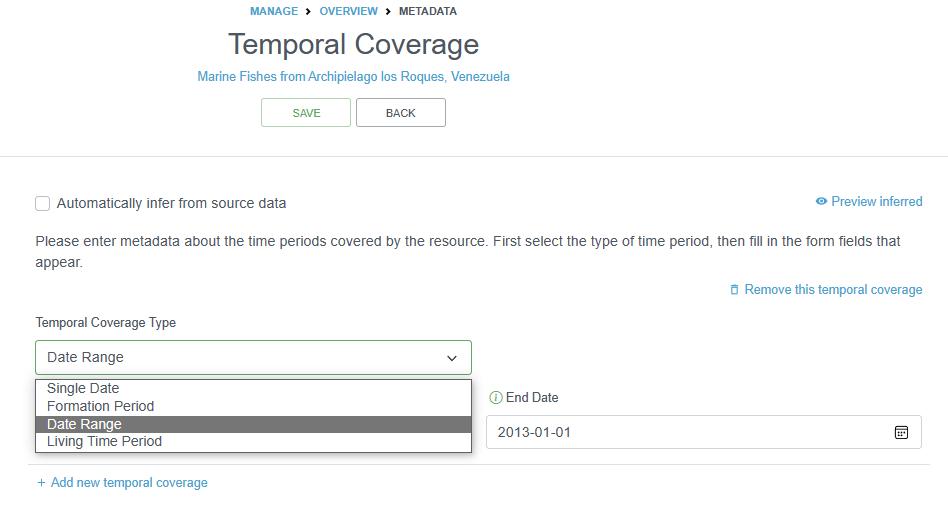
2.4.4.1.5.3 Temporal Coverage
The temporal coverage will be a date range, which can easily be documented. If it is a single date, the start and end date will be the same. The information added here can be used as a quality check for the actual dates in the datasets.
You can also document the Formation Period or the Living Time Period in this section for specimens that may not have been alive during the collection period, or to indicate the time during which the collection occurred.
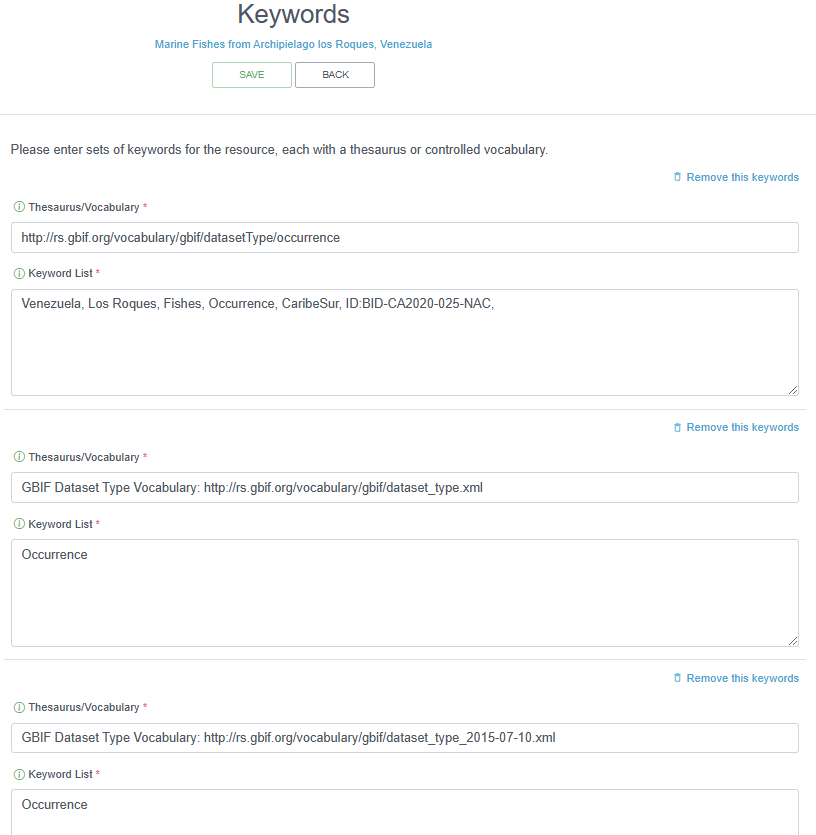
2.4.4.1.6 Keywords
Relevant keywords facilitate the discovery of a dataset. An indication of the represented functional groups can help in a general search (e.g. plankton, benthos, zooplankton, phytoplankton, macrobenthos, meiobenthos …). Assigned keywords can be related to taxonomy, habitat, geography or relevant keywords extracted from thesauri such as the ASFA thesaurus, the CAB thesaurus or GCMD keywords.
As taxonomy and geography are already covered in previous sections, there is no need to repeat related keywords here. Please consult your data provider which (relevant) keywords can be assigned.
2.4.4.1.7 Project
If the dataset in this resource is produced under a certain project, the metadata on this project can be documented here. Part of the information entered here, can partly overlap with information given in other sections of the metadata (e.g. study area description can have lot of parallel with the geographic coverage section). Personnel involved in the project can be documented or repeated here as well. This is not a problem.
2.4.4.1.8 Sampling Methods
The EML can contain descriptions of the sampling and data processing methods. Study extent can be documented here as well to report a more specific geographic area as well as the sampling frequency. Descriptions of sampling procedures, quality control, and steps (sample or data processing) can be given in the same way as the methods section of a scientific paper.
Note that OBIS best practice is to add sampling facts to the extended MeasurementorFact extension, linked to the sampling events in the Event core via eventID.
2.4.4.1.9 Citations
The dataset citation allows users to properly cite your dataset in further publications or other uses of the data. When users download datasets from the OBIS download function, a list of the dataset citations packaged with the data in a zipped file is provided.
A dataset citation is different from the data source citation (in case the data is digitized from a publication), and these references can be added to the additional metadata (see bibliography below). A dataset citation can have the same format of a journal article citation, and should include the authors (contact, creator, principle investigator, data managers, custodians, collectors…), the title of the dataset, the name of the data publisher (or custodian institute), and the access point URL to the resource.
GBIF’s IPT has an auto-generation - Turn On/Off - tool to let the IPT auto-generate the resource citation for you. The citation includes a version number, which is especially important for datasets that are continuously updated. The dataset citation can also include a Citation Identifier - a DOI, URI, or other persistent identifier that resolves to an online dataset web page.
The OBIS node data managers should try to implement a certain degree of format standardization for the dataset citations. The IPT provides an option to auto-generate a citation based on the EML and is formatted as follows: {dataset.authors} ({dataset.pubDate}) {dataset.title}. [Version {dataset.version}]. {organization.title}. {dataset.type} Dataset {dataset.doi}, {dataset.url}
2.4.4.1.10 Bibliography
The EML can include the citation of the publications that are related to the described dataset. They can describe the dataset, be based on the dataset or be used in this dataset. Publications can be scientific papers, reports, PhD or master theses. If available, the citation should include the DOI at the end.
This overview will contribute to a better understanding of the data as these publications can hold important additional information on the data and how they were acquired.
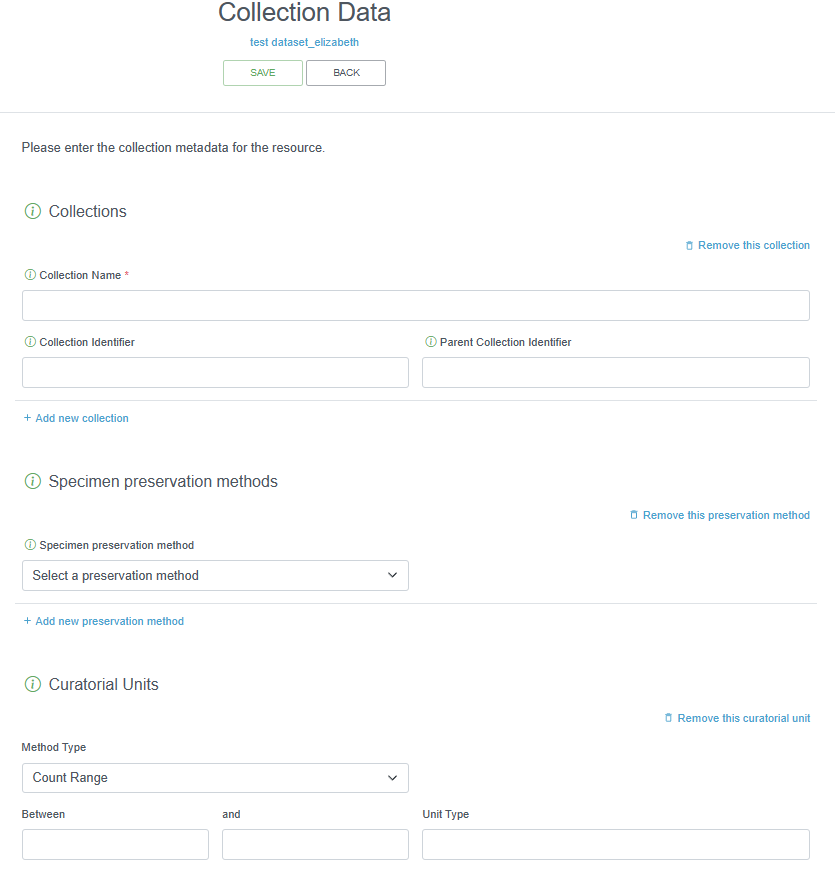
2.4.4.1.11 Collection Data
This IPT section should only be filled out if there are specimens held in a museum. If relevant, it is strongly recommended that this information is supplied by the data provider or left blank. The collection name, specimen preservation method, and curatorial units should be provided, as applicable.
2.4.4.1.12 External Links
This section can include URLs to the resource homepage, to download or find additional information. You can also provide links to your resource if it is hosted elsewhere in different formats.
Links to the online dataset on the OBIS website can be added once the data is available there. For these OBIS links, the required fields should be completed as follows:
- Name: online dataset
- Character set: UTF-8
- Data format: html
If other links are added, then the data format for web-based data is ‘html’. If the link refers to a file, the data format of the file will need to be added (e.g. .xlsx, .pdf …). The character set for all Darwin Core files is UTF-8, whereas for other web pages this can vary, so you may need to confirm.
2.4.4.1.13 Additional Metadata
Any remaining information that could not be catalogued under any of the other metadata, can be mentioned here. This may include logos, purpose of the dataset, a description of how the dataset will be maintained, etc.
2.5 OBIS nodes
Note the OBIS node TOR and system architecture is currently under review and will be updated after the 2023 Steering Group meeting. The information below may change.
OBIS Nodes are either national projects, programmes, institutes, or organizations, National Ocean Data Centers or regional or international projects, programmes and institutions or organizations that carry out data management functions.
OBIS nodes are responsible for representing all aspects of OBIS within a particular region or taxonomic domain. Additional responsibilities include:
- Establishing relationships with key data providers within their geographical (or taxonomic) area of responsibility
- Bringing data and corresponding metadata into the global database to be shared with the OBIS community
- Responsibility for all aspects of the data
- Gaining permission to providing access to the data
- Ensuring a certain level of data quality
- Transfer of these datasets to the global OBIS database
- Provide support for the full implementation of OBIS worldwide by serving on the IODE Steering Group for OBIS and any relevant Task Teams or ad hoc project teams
- Each node may also maintain a data presence on the Internet representing their specific area of responsibility
2.5.1 Terms of Reference of OBIS nodes
Data Responsibilities
- Receiving or harvesting marine biodiversity data (and metadata) from national, regional, and international programs, and the scientific community at large, and from Tier III nodes by Tier II nodes, and from Tier II nodes by Tier I nodes
- Perform data validation (using standards, tools, and best practices), as described in the OBIS manual (Tier II)
- Reporting the results of quality control directly to data collectors/originator (or Tier III node) as part of the quality assurance activity
- Making data (and metadata) available to OBIS using agreed upon standards and formats which are described in the OBIS Manual (Tier II), making data available to Tier II nodes (Tier III)
- Control data access, terms of use and sharing policies
- Comply with the IOC/OBIS data policy for using and sharing OBIS data
- Contribute to the development of standards and best practices in OBIS (recommended)
- Contribute to the development of open-source tools in OBIS (recommended)
- Ensuring the long-term preservation of the data, metadata and associated information required for correct interpretation of the data (including version-control) (recommended)
- Build customized data portals (optional)
Administration Responsibilities
- Become a member of the IODE steering group for OBIS, attend the SG-OBIS annual meeting and report on node activities
- Provide indicators on up-time, responsiveness, and data processed by nodes and present a report to SG-OBIS
- Customer support (data queries, analyses, feedback)
- Outreach and Capacity Building (i.e., providing expertise, training and support in data management, technologies, standards and best practices)
- Engage in stakeholder groups (recommended)
2.5.2 How to become an OBIS node
OBIS nodes now operate under the IODE network as either National Oceanographic Data Centres (NODCs) or Associate Data Unites (ADUs). Prospective nodes are required to apply to the IODE for membership.
The procedure to become an OBIS node is as follows:
- If you are an existing NODC (within the IODE network) and the OBIS node activities fall under the activities of the NODC:
- Send a letter expressing your interest to become an OBIS node (including contact information of the OBIS node manager, and geographical/thematic scope of your OBIS node)
- If you are not an existing NODC:
- Email your application form to become an IODE Associate Data Unit (ADU), with a specific role as OBIS node. Applications for ADU membership in OBIS shall be reviewed by the IODE Officers in consultation with the IODE Steering Group for OBIS.
2.5.3 OBIS Node Health Status Check and Transition Strategy
OBIS nodes should operate under IODE as either IODE/ADU or IODE/NODC. As such OBIS nodes are a member of the IODE network.
The IODE Steering Group (SG) for OBIS evaluates the health status of OBIS nodes at each annual SG meeting, and considers an OBIS node as inactive when it meets any of the following conditions:
- The OBIS node manager recurrently fails to answer the communications from the project manager or the SG co-chairs in the last 12 months
- The OBIS node manager or a representative fails to attend (personally or virtually) the last 2 SG meetings without any written reason
- The OBIS node does not have an IPT
- The OBIS node has an IPT, but it has not been running for the last 12 months
- The datasets in the OBIS node’s IPT have been removed and not restored in the last 12 months (without any explanation)
- The OBIS node has not provided new data for the last 2 years
The OBIS Secretariat prepares a health status check report of each OBIS node based on the six items above and informs the OBIS node manager on their status 3 months before the SG meeting. At the SG meeting, the SG-OBIS co-chair will present the results of the OBIS nodes health status check report including a listing of the inactive OBIS nodes. The SG-OBIS members representing active OBIS Nodes will make one of the following decisions:
- Request the inactive OBIS node to submit a plan with actions, deliverables and times to improve their performance, within 3 months, to the OBIS Secretariat. This plan is reviewed and accepted by the OBIS-Executive Committee Or
- Provide a recommendation to the IOC Committee on IODE to remove the OBIS node from the IODE network.
In either case, the OBIS Secretariat will inform the OBIS node manager of the SG-OBIS decision, with a copy to the IODE officers and the IODE national coordinator for data management of the country concerned.
The IODE Committee is requested to consider the recommendation from the OBIS Steering Group and it may either accept the recommendation or request the inactive OBIS node to submit an action plan (option 1).
When the inactive OBIS node is removed from the IODE network, the SG-OBIS will ask whether another OBIS node is interested in taking over the responsibilities of the removed OBIS node, until a new OBIS node in the country/region is established.